This protocol describes how to sequence using a MinION the DNA extracted using the Beer DNA extraction protocol.
The protocol is in 3 parts:
- Preparation of the MinION with flow cell and software preparation
- Library preparation, i.e. prepare the DNA for sequencing
- Loading the DNA into the flow cell
- Sequencing using the nanopore technology
To prepare before
For this protocol, we will need ~400 ng DNA.
For this protocol, we will need ~200 ng DNA.
Consumables
MinION preparation
Library preparation
- 400ng DNA solved in 7.5µl nuclease-free water, extracted given the DNA extraction protocol
- 200ng DNA solved in 7.5µl nuclease-free water, extracted given the DNA extraction protocol
- Rapid Sequencing Kit including
- 2.5 µl Fragmentation Mix (FRA - amber cap)
- 1 µl Rapid Adapter (RAP - green cap)
- 1.25 µl Fragmentation Mix (FRA - amber cap)
- 0.5 µl Rapid Adapter (RAP - green cap)
- Nuclease-free water (e.g. ThermoFisher, cat #AM9937)
- 0.2 mL PCR tubes
- Ice
Loading the DNA into the flow cell
- MinION Flow Cell
- Flongle Flow Cell
Rapid Sequencing Kit including
- 30 µl Flush Tether (FLT - purple stripe cap)
- 34 μl Sequencing Buffer (SQB - red cap)
- 1 tube of Flush Buffer (FB - blue cap)
- 25.5 μl Loading Beads (LB - pink cap)
- 3 µl Flush Tether (FLT - purple stripe cap)
- 13.5 μl Sequencing Buffer (SQB - red cap)
- 117 μl of Flush Buffer (FB - blue cap)
- 11 μl Loading Beads (LB - pink cap)
- Nuclease-free water (e.g. ThermoFisher, cat #AM9937)
- 1.5 ml Eppendorf DNA tubes
- MinIon device including Flow cell
- Ice
Material
MinION preparation
- Computer with at least 8GB RAM and 4 CPU
- MinION
Library preparation
- Thermocycler at 30° C and 80° C
- P2 pipette and tips
- P10 pipette and tips
- Centrifuge
Loading the DNA into the flow cell
- Computer with at least 8GB RAM and 4 CPU
- MinION with the flow cell
- P100 pipette and tips
- P1000 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Vortex
- Centrifuge
Sequencing
- Computer with at least 8GB RAM and 4 CPU
- MinION with the loaded flow cell
MinION preparation
Needed consumables:
- MinION Flow Cell
- Flongle Flow Cell
Needed material:
- Computer with at least 8GB RAM and 4 CPU
- MinION
Installation and preparation of the MinKNOW software
MinKNOW software controls the small sequencing device (MinION) and collects sequencing data in real-time. The software has a graphical user interface for configuring and running the sequencing process. Furthermore, it has an integrated base-calling software to convert the raw nanopore signals to the strings of nucleotides, in fastq file format.
- Create an account on the Nanopore Community
- Search for “MinION Software” on the downloads page of Nanopore
- Select the operating system of the computer and download it
- Follow the installation instructions
-
Launch the software called MinKNOW
On Linux
- Open a terminal
- Run
/opt/ui/MinKNOW
Preparation of the MinION
-
Connect the MinION to the computer via USB
We should see a light and hear the fan on the MinION once successfully plugged in.
-
Launch the MinKNOW software
-
Open the MinION lid
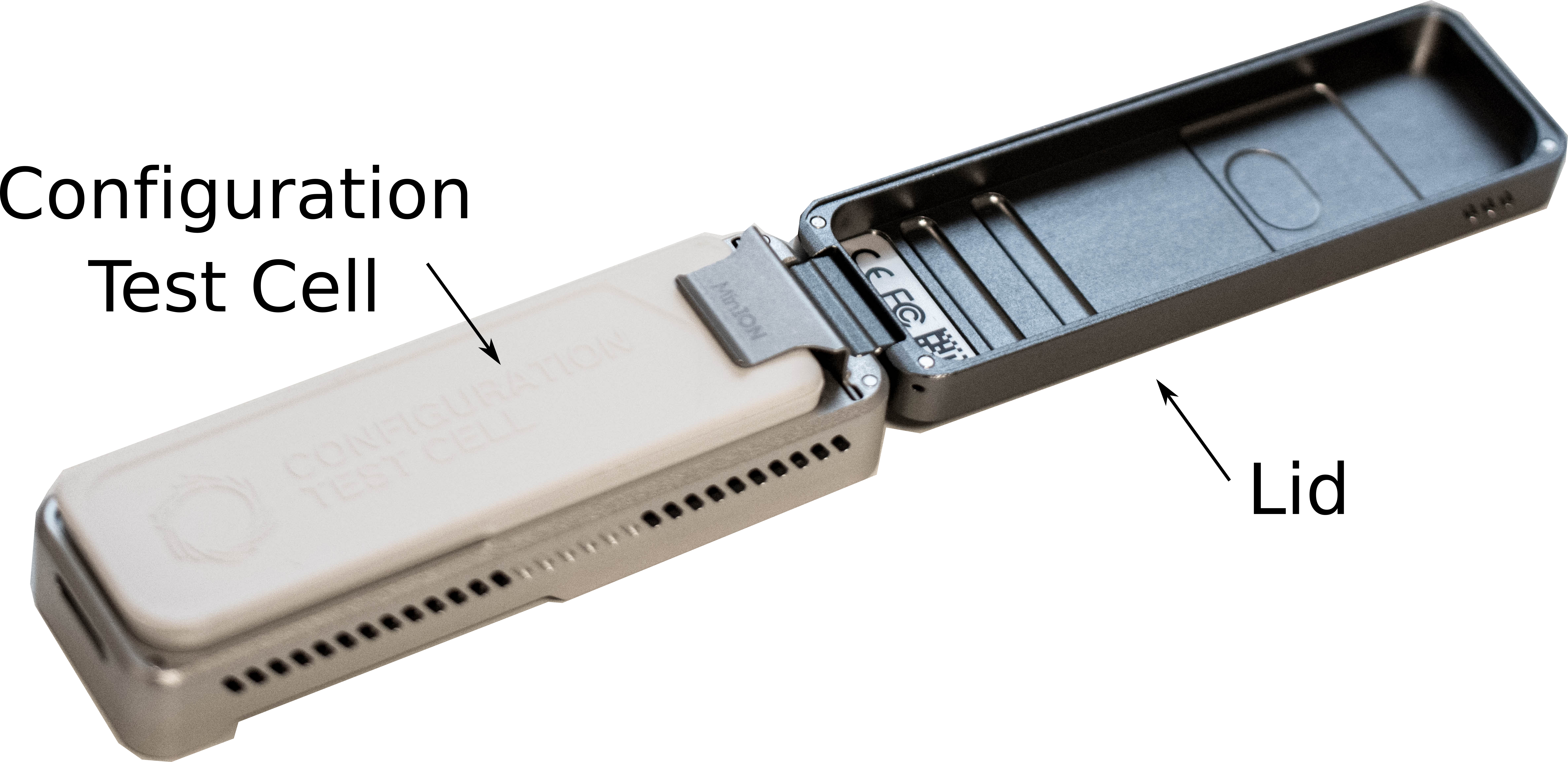
-
Remove the Configuration Test Cell
-
Add a flow cell
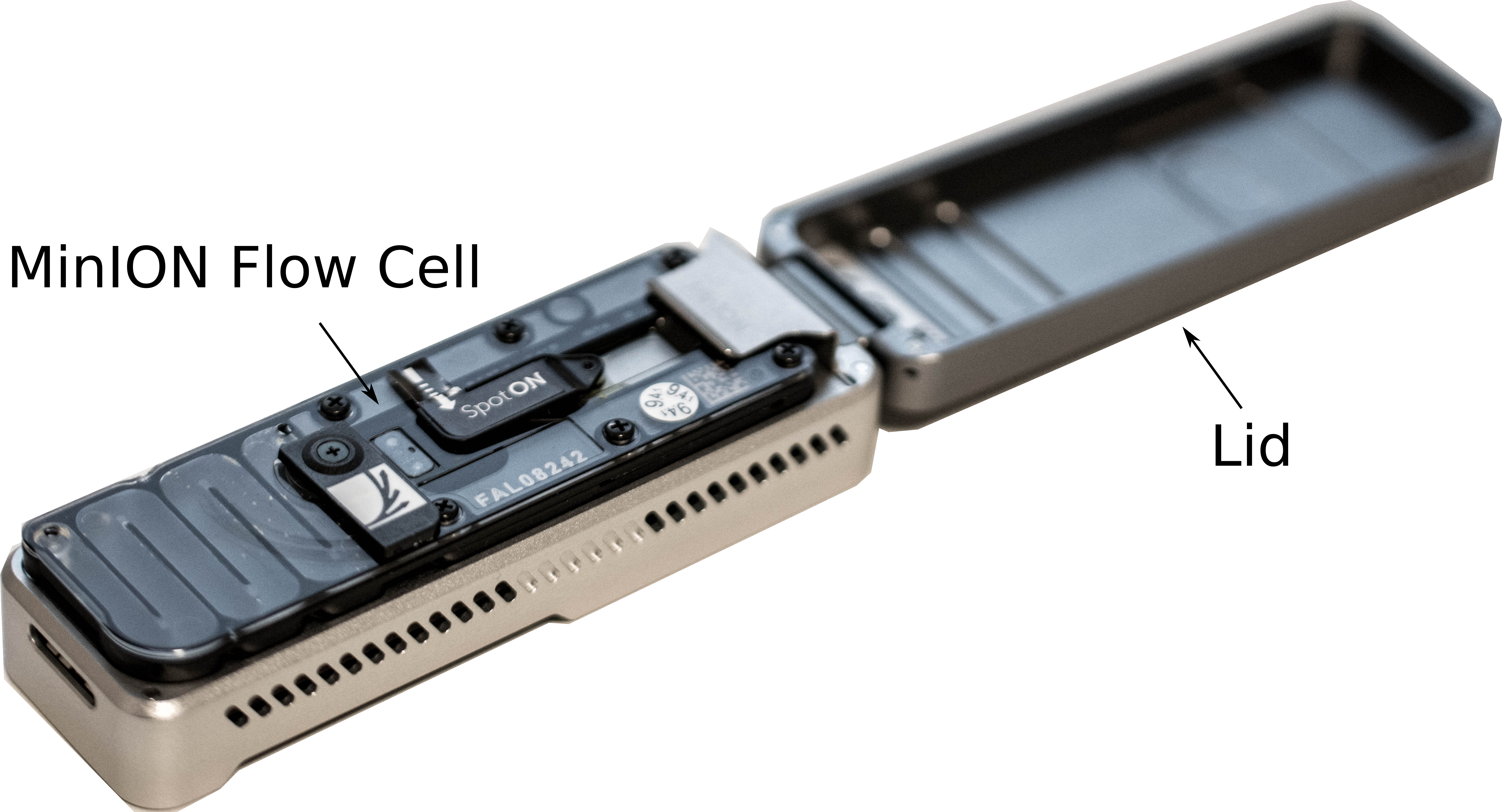
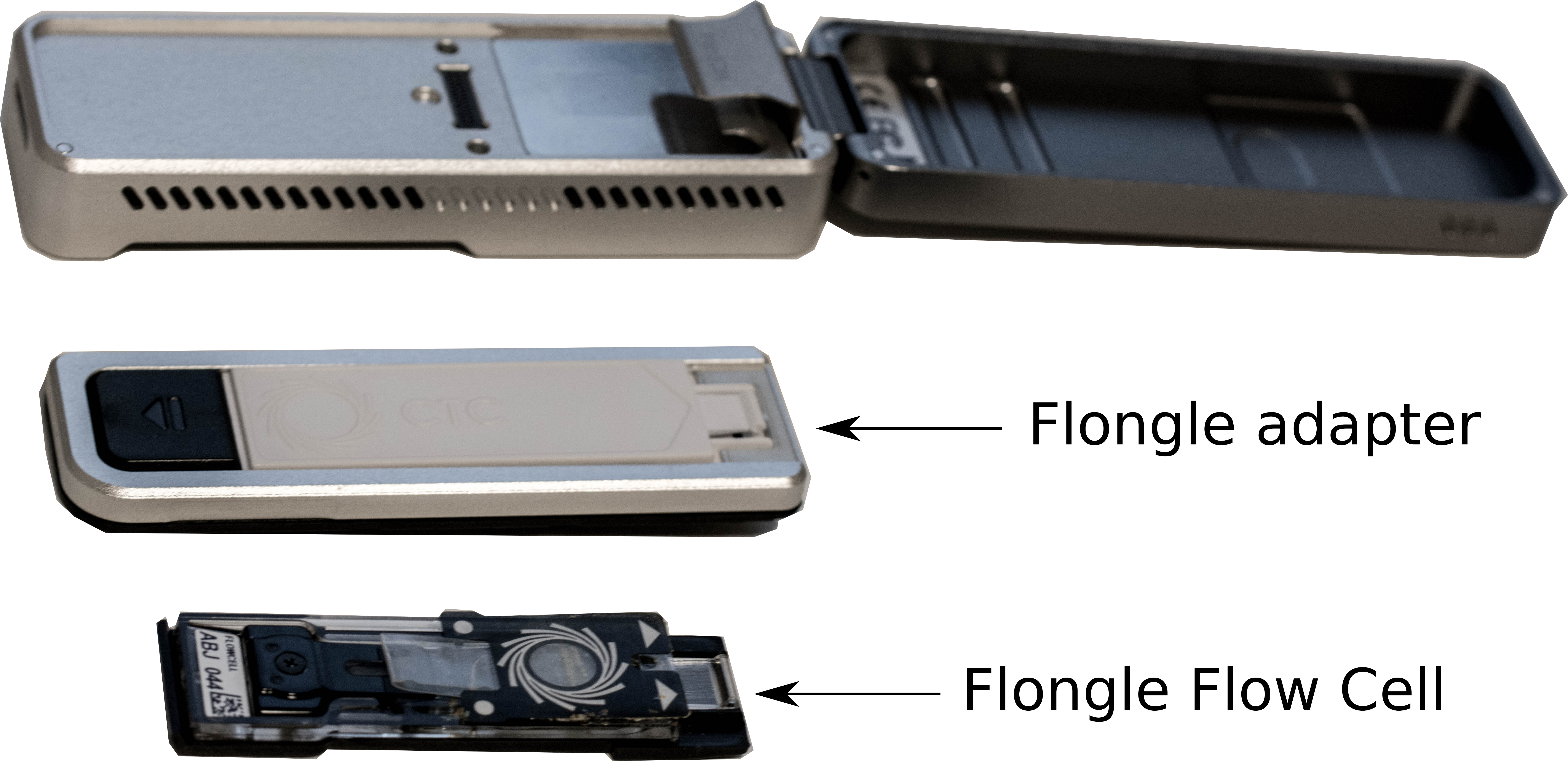
- Remove the CTC on the adapter
- Add the Flongle Flow Cell on the adapter
- Add the adapter with the Flongle Flow Cell on the MinION
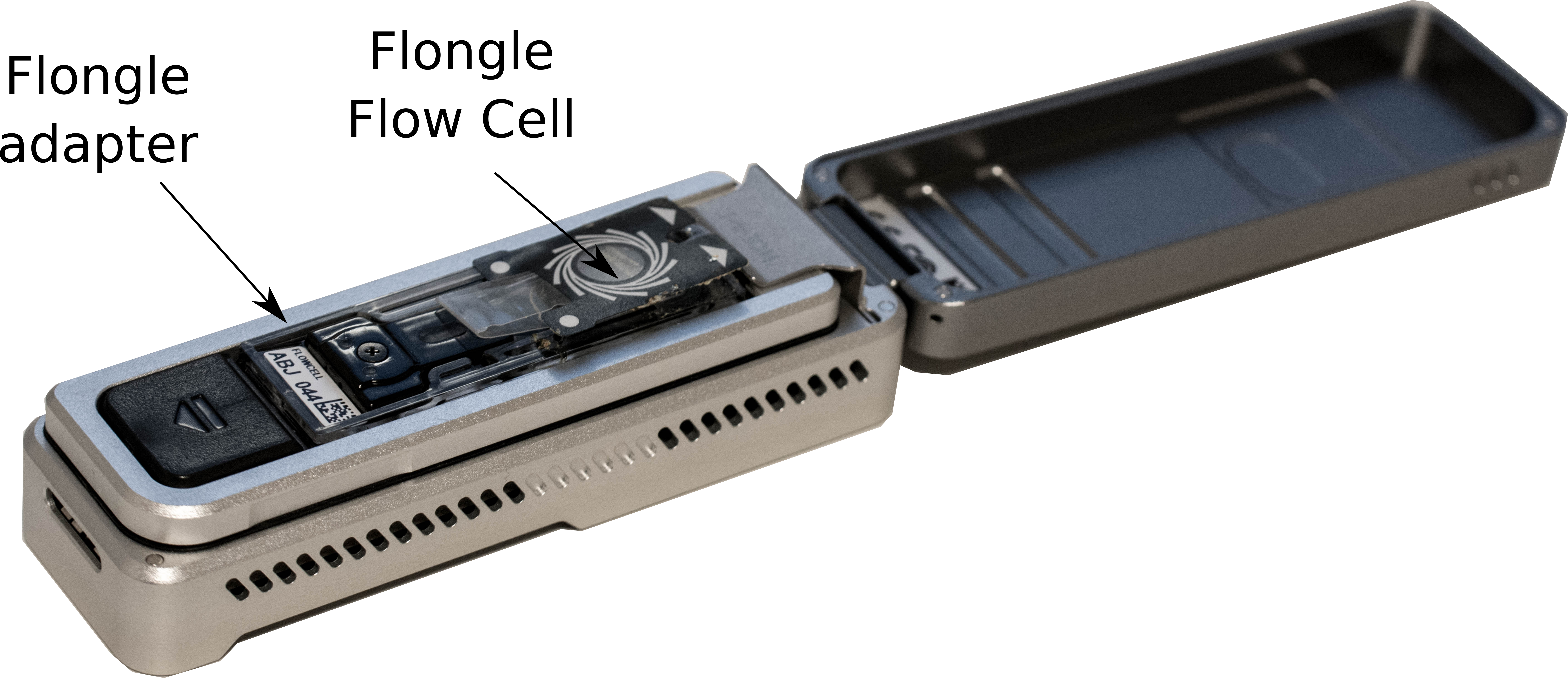
-
Click on the flow cell in MinKNOW software
With Flongle, we need to assign the Flow Cell ID manually. The ID is shown in the Flongle box in blue.
-
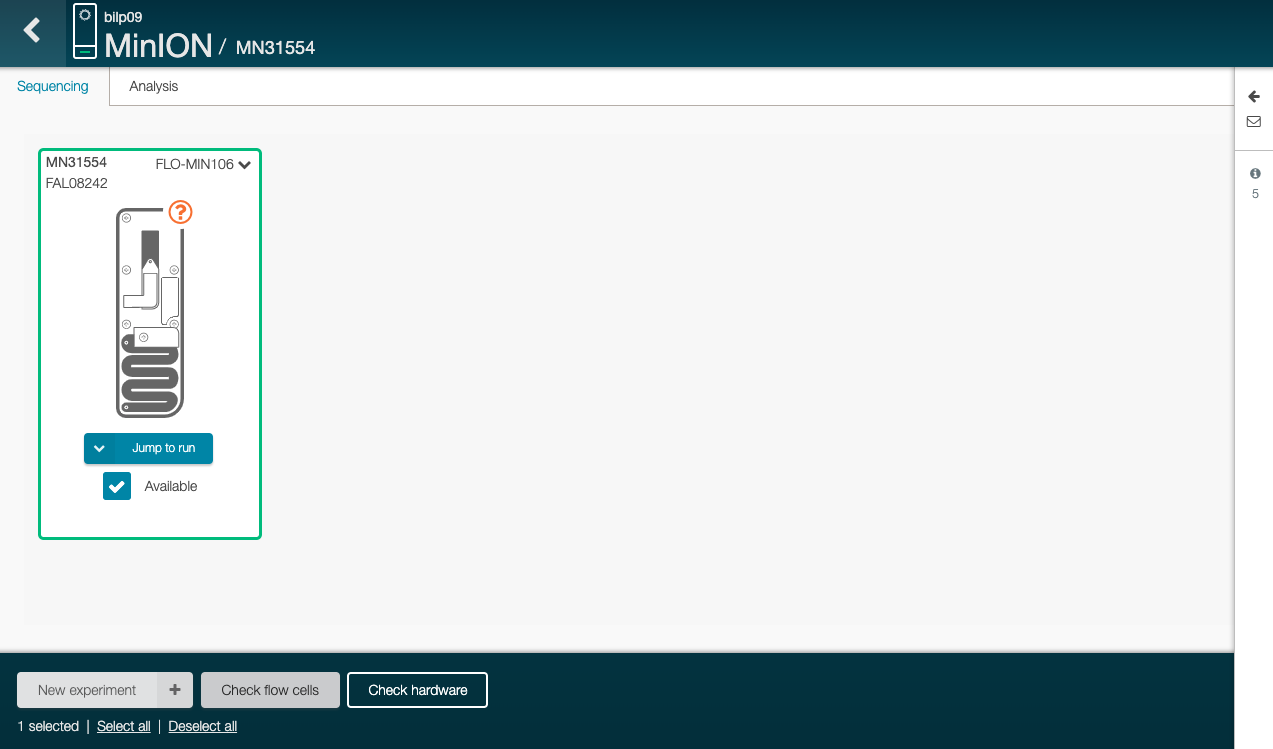
Click on “Check flow cells” (on the bottom)
The test assess the number of nanopores that are available in the flow cell.
-
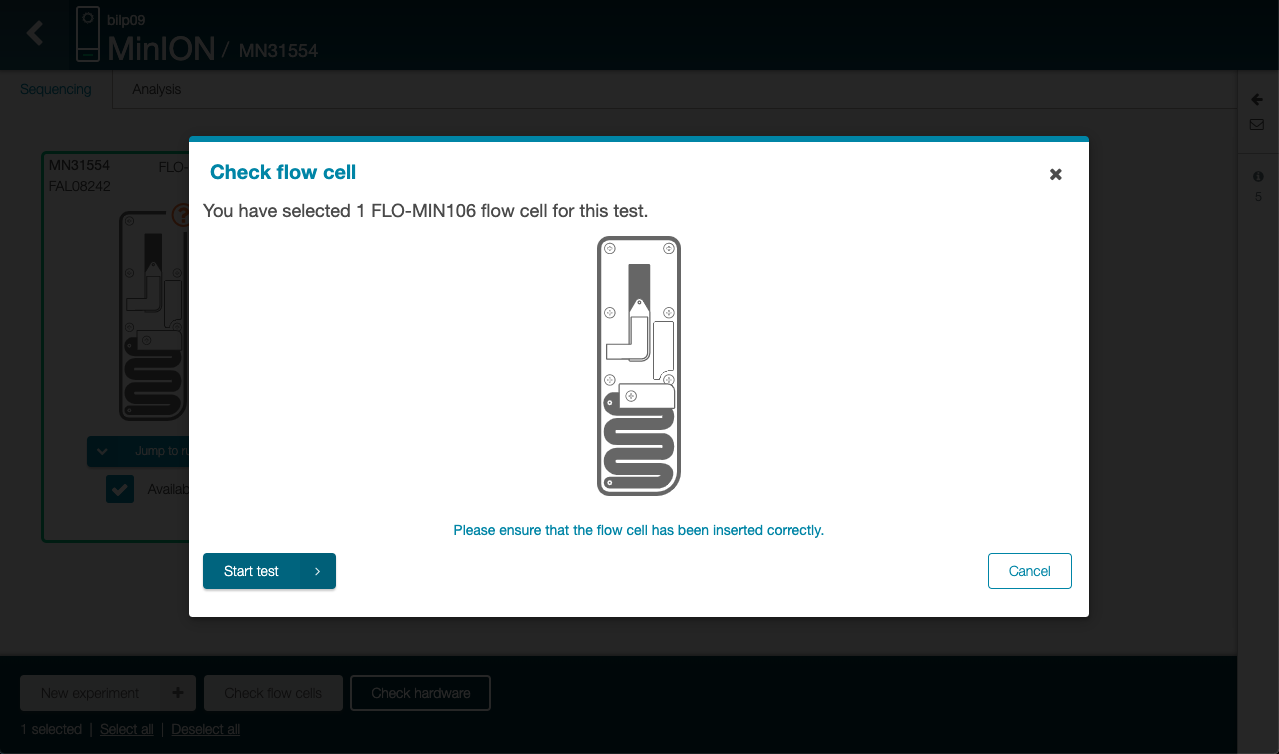
Click on “Start test” in the new
-
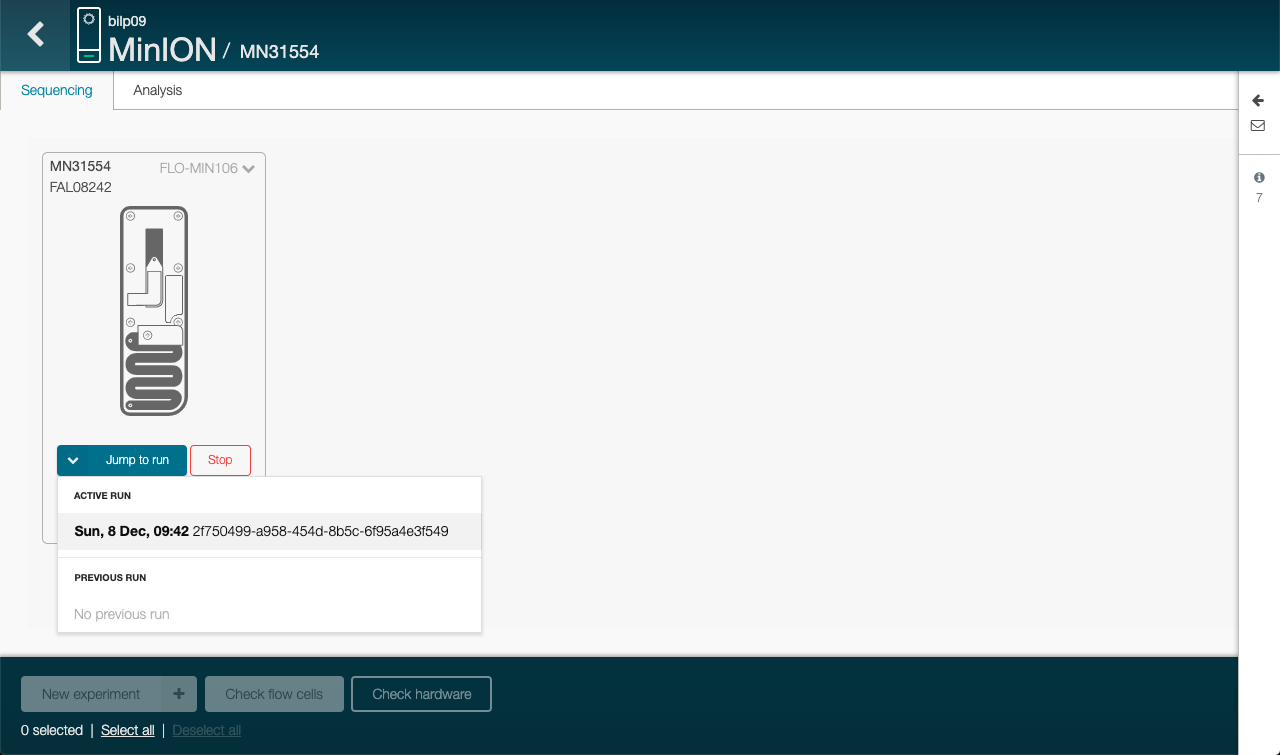
Click on Jump to run and then the active run, on the welcome page of MinKNOW
-
Wait for the tests to be done
The flow cell is first getting to 37°C and then each nanopores in the flow cell are checked.
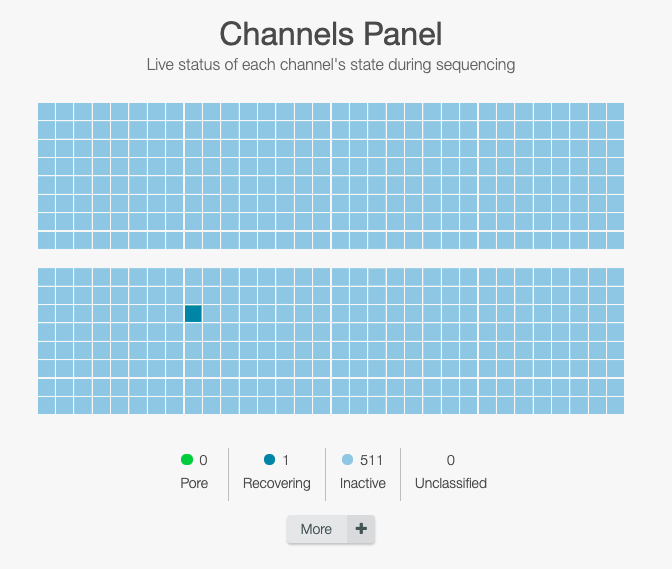
Due to sequencing runs and time, the nanopore complexes tend to be deteriorated and become dead, and could not read the DNA strand.
-
Click on the right side under message to check how many pores are available for sequencing
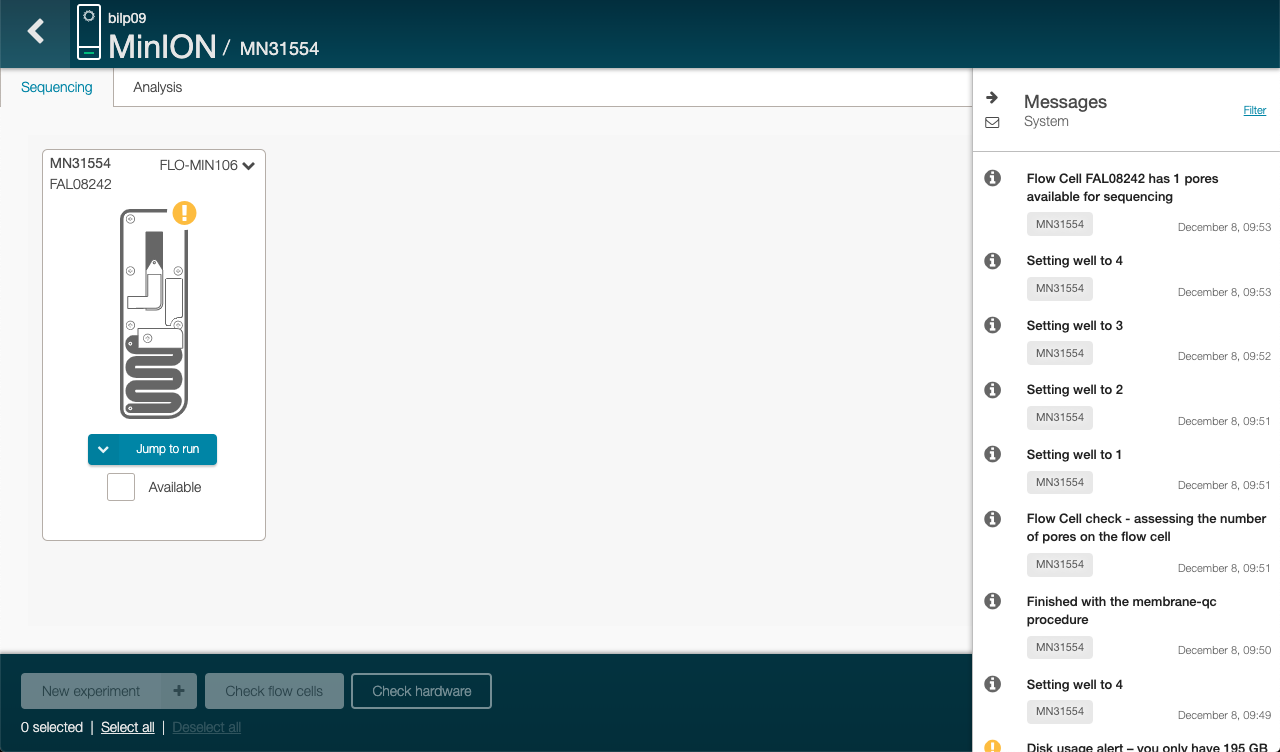
To run sequencing, we need at least 800 nanopores.
To run sequencing, we need at least 60 nanopores for FLO-FLG001 and 30 for FLO-FLGOP1.
Library preparation
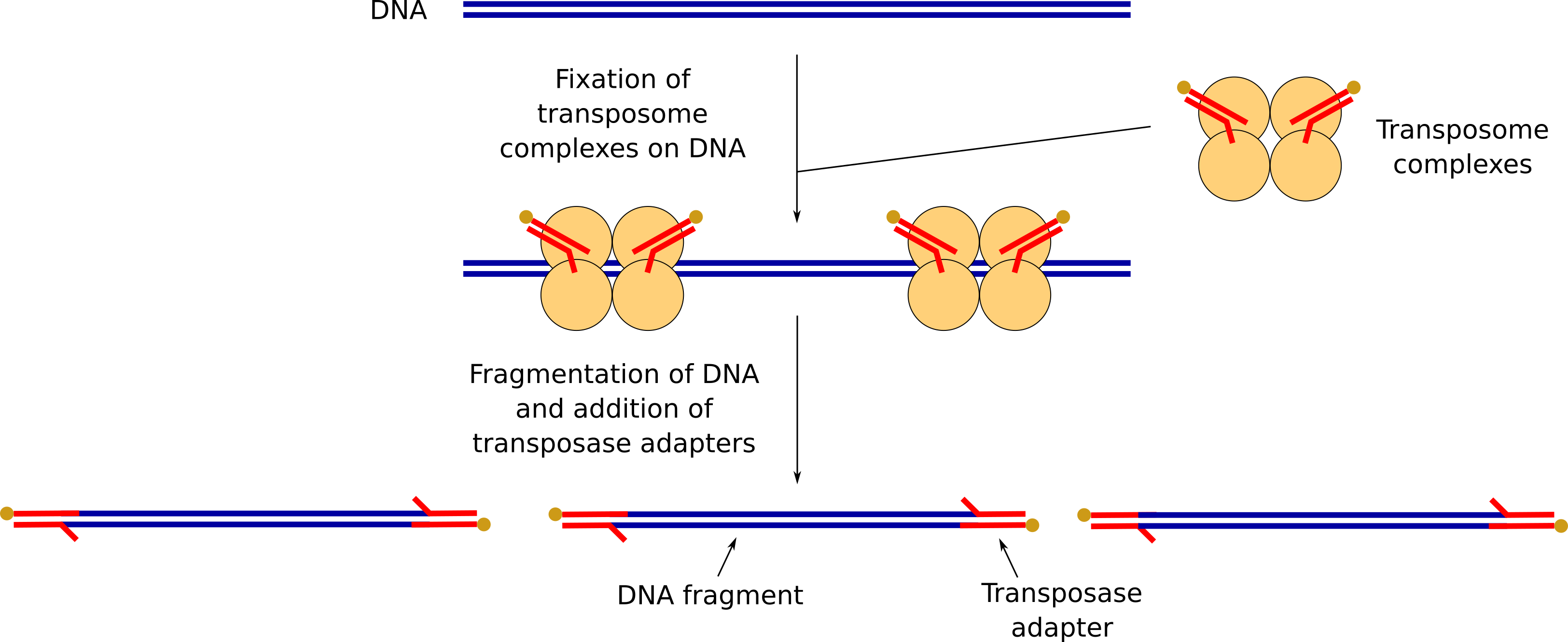
We need now to prepare the DNA to attach sequencing adapters that will lead the DNA fragments through the nanopores. This step is called “Library preparation” (a library being a collection of randomly sized DNA represented the sample, here the beer yeast DNA) and consist of:
- Fragmentation of our DNA and addition some transposase adapters
- Attachement of sequencing adapters on the top of the transposase adapters
Needed consumables:
- 400ng DNA solved in 7.5µl nuclease-free water, extracted given the DNA extraction protocol
- 200ng DNA solved in 7.5µl nuclease-free water, extracted given the DNA extraction protocol
- Rapid Sequencing Kit including
- 2.5 µl Fragmentation Mix (FRA - amber cap)
- 1 µl Rapid Adapter (RAP - green cap)
- 1.25 µl Fragmentation Mix (FRA - amber cap)
- 0.5 µl Rapid Adapter (RAP - green cap)
- Nuclease-free water (e.g. ThermoFisher, cat #AM9937)
- 0.2 mL PCR tubes
- Ice
Needed material:
- Thermocycler at 30° C and 80° C
- P2 pipette and tips
- P10 pipette and tips
- Centrifuge
DNA fragmentation and addition of transposase adapters
- Prepare the sequencing kit (FRA & RAP)
- Thaw at room temperature
- Briefly spin down each reagent briefly by vortexing
- Mix well by pipetting (up and down) each reagent
- Keep all the kit components on ice once thawed
Before using each reagent, repeat the mixing step
- Prepare the DNA
- Transfer the ~400 ng DNA into a new tube
- Transfer the ~200 ng DNA into a new tube
-
Adjust the volume to 7.5 μl with nuclease-free water
“Normal” water contains nucleases which may degrade DNA
-
Adjust the volume to 3.75 μl with nuclease-free water
“Normal” water contains nucleases which may degrade DNA
- Mix by flicking the tube
- Spinning down briefly in a centrifuge, e.g. at 10 sec at 8000 RPM (use the “short” button on your centrifuge if available)
-
Prepare a PCR tube with the 7.5 µl extracted DNA and 2.5 µl FRA
FRA contains transposome complexes with transposase adapters. The transposome complexes will fix at different positions on the DNA.
-
Prepare a PCR tube with the 3.75 µl extracted DNA and 1.25 µl FRA
FRA contains transposome complexes with transposase adapters. The transposome complexes will fix at different positions on the DNA.
- Mix gently by flicking the tube, and spin down
-
Put the tubes into a thermocycler for 1 min at 30°C and then 1 min at 80°C
The DNA will be fragmented where the transposome complexes are fixed and transposase adapters will be added there.
- Put the tube on ice until next step
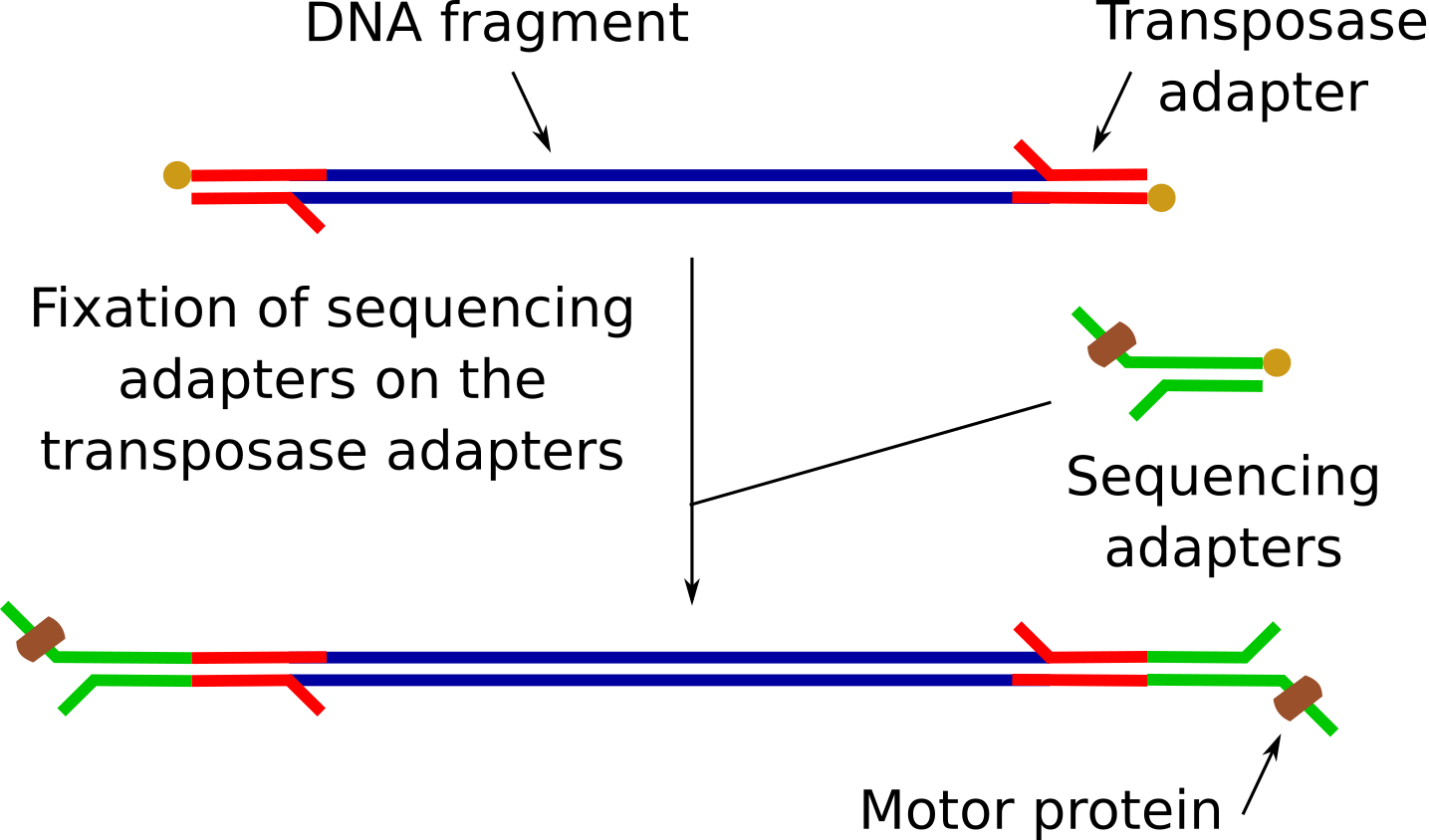
Sequencing adapter attachment
-
Add 1 µl of RAP into the tube
RAP contains sequencing adapters that are ligated to the DNA on the transposase adapters.
These sequencing adapters have the motor proteins, needed for the sequencing to guide the DNA fragments through the nanopores.
-
Add 0.5 µl of RAP into the tube
RAP contains sequencing adapters that are ligated to the DNA on the transposase adapters.
These sequencing adapters have the motor proteins, needed for the sequencing to guide the DNA fragments through the nanopores.
- Mix gently by flicking the tube, and spin down
- Incubate the reaction for 5 min at room temperature
- Store the library on ice until ready to load
Priming and loading the flow cell
The DNA is ready, so we can now load it into the flow cell to start the sequencing. Before, we need to prime the flow cell.
Needed consumables:
- MinION Flow Cell
- Flongle Flow Cell
Rapid Sequencing Kit including
- 30 µl Flush Tether (FLT - purple stripe cap)
- 34 μl Sequencing Buffer (SQB - red cap)
- 1 tube of Flush Buffer (FB - blue cap)
- 25.5 μl Loading Beads (LB - pink cap)
- 3 µl Flush Tether (FLT - purple stripe cap)
- 13.5 μl Sequencing Buffer (SQB - red cap)
- 117 μl of Flush Buffer (FB - blue cap)
- 11 μl Loading Beads (LB - pink cap)
- Nuclease-free water (e.g. ThermoFisher, cat #AM9937)
- 1.5 ml Eppendorf DNA tubes
- MinIon device including Flow cell
- Ice
Needed material:
- Computer with at least 8GB RAM and 4 CPU
- MinION with the flow cell
- P100 pipette and tips
- P1000 pipette and tips
- P20 pipette and tips
- P10 pipette and tips
- Vortex
- Centrifuge
- Prepare the reagents (SQB, LB, FLT and FB)
- Thaw at room temperature
- Mix the Sequencing Buffer (SQB) and Flush Buffer (FB) tubes by vortexing, spin down and return to ice
- Spin down the Flush Tether (FLT) tube, mix by pipetting, and return to ice
-
Prepare MinION for loading
-
Slide the priming port cover clockwise to make the priming port visible

- Remove possible bubbles
- Set of P1000 pipette to 200 µl
- Insert the tip into the priming port
-
Turn the wheel of the pipette until 220-230 µl
Removing more than 30 µl will damage the pores in the array because they need to be covered by the buffer at all times
- Check visually that the buffer is continuous from the priming port to the sensor array via the inlet channel
-
- Prepare the priming mix
- Add 30 µl of Flush Tether (FLT) directly to the Flush Buffer (FB) tube
- Mix 3 µl of Flush Tether (FLT) with 117 µl of Flush Buffer (FB) in a 1.5 ml Eppendorf DNA tube
- Mix by pipetting up and down
The priming mix includes chemicals that improve the rate of sequencing.
- Remove the seal table from the Flongle flow cell to expose the sample port
- Lift up the seal tab
- Pull the seal tab to open access to the sample port
- Hold the seal tab open by using adhesive on the tab to stick to the lid

- Load 800 μl of the priming mix via the priming port, avoiding introduction of air bubbles
- Load the priming mix via the sample port
- Load the primix mix into a P200 pipettte
- Place the P200 pipette tip inside the sample port
- Slowly dispense the priming fluid by twisting the pipette plunger down
Make sure there is no air gap in the sample port or the pipette tip
- Wait for 5 minutes
- Prepare library for loading by mixing in a new tube
- 34 μl Sequencing Buffer (SQB)
- 13.5 μl Sequencing Buffer (SQB)
-
25.5 μl Loading Beads (LB) (mix before adding)
The loading beads bring the DNA fragments closer to the nanopores.
-
11 μl Loading Beads (LB) (mix before adding)
The loading beads bring the DNA fragments closer to the nanopores.
- 4.5 μl nuclease-free water
- 11 μl DNA library
- 5.5 μl DNA library
-
Load the library mix into the flow cell
-
Gently lift the SpotON port cover to make the SpotON port accessible

-
Load 200 μl of the priming mix into the priming port (not the SpotON port), avoiding introduction of air bubbles
It clears any blockages near the SpotON port.
- Mix the prepared library gently by pipetting up and down prior to loading
-
Add 75 μl of the library preparation into the SpotON port drop by drop
Make sure that each drop flows into the port before adding the next
- Gently replace the SpotON port cover
- Making sure the bung enters the SpotON port
- Close the priming port
- Load the library mix into a P100 pipette
- Place the P100 tip inside the sample port
- Slowly dispense the priming fluid by twisting the pipette plunger down
- Stick the adhesive tape to the sample port
- Replace the seal tab
-
- Close the MinION lid
Sequencing
We can now start the sequencing itself.
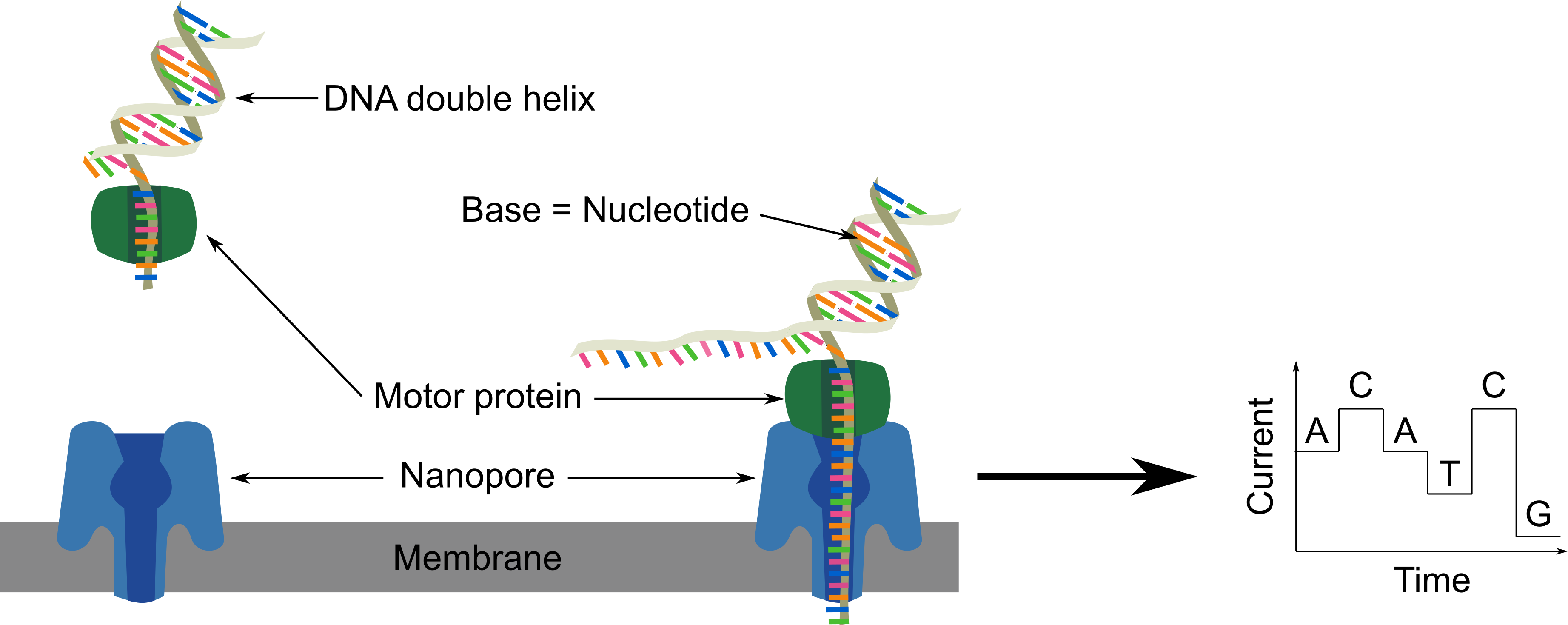
The DNA fragments going through nanopores modify the electric current. The difference of current intensity depends on the nucleotide (A, T, C, G). This information is used to directly read the nucleotide sequence of a DNA fragment.
The DNA fragments in the library mix are directed toward the nanopores on the flowcell. The motor protein on the DNA fragment are fixed to the nanopore, open the double strand DNA, direct one strand through the nanopore in which bases are read one by one. Once the full fragment is read, another one will be read such as at the end all DNA fragments are read.
To read the differences of current intensity and transform it into a useful nucleotide sequence, we need to use the MinKNOW software, which will collect raw data from the MinION and convert it into sequences.
-
Launch the MinKNOW software

-
Select on the flow cell

-
Click on “New Experiment” on the bottom left
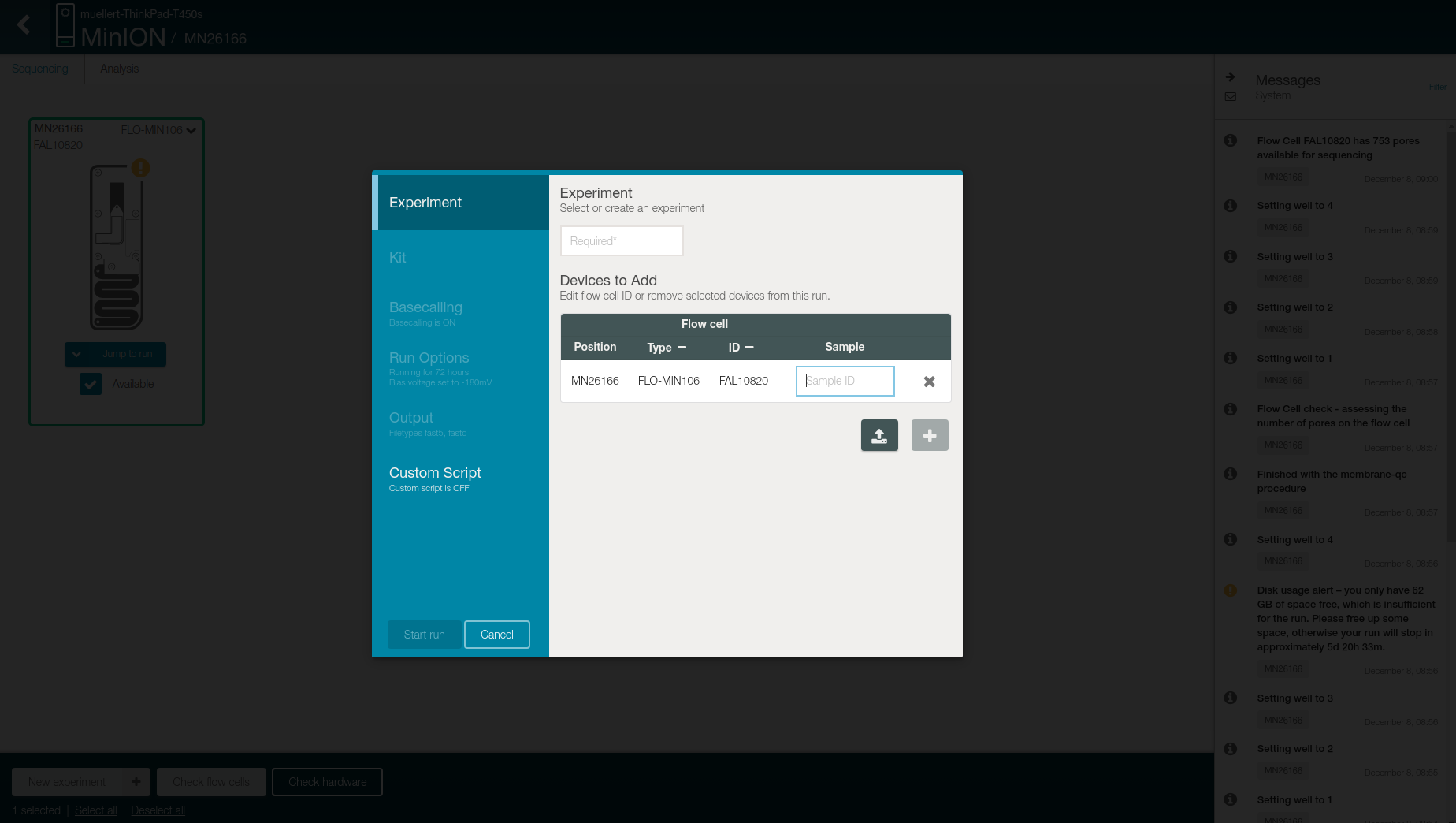
-
Add the name of the beer as experiment name in the first box and the date in the sample box
-
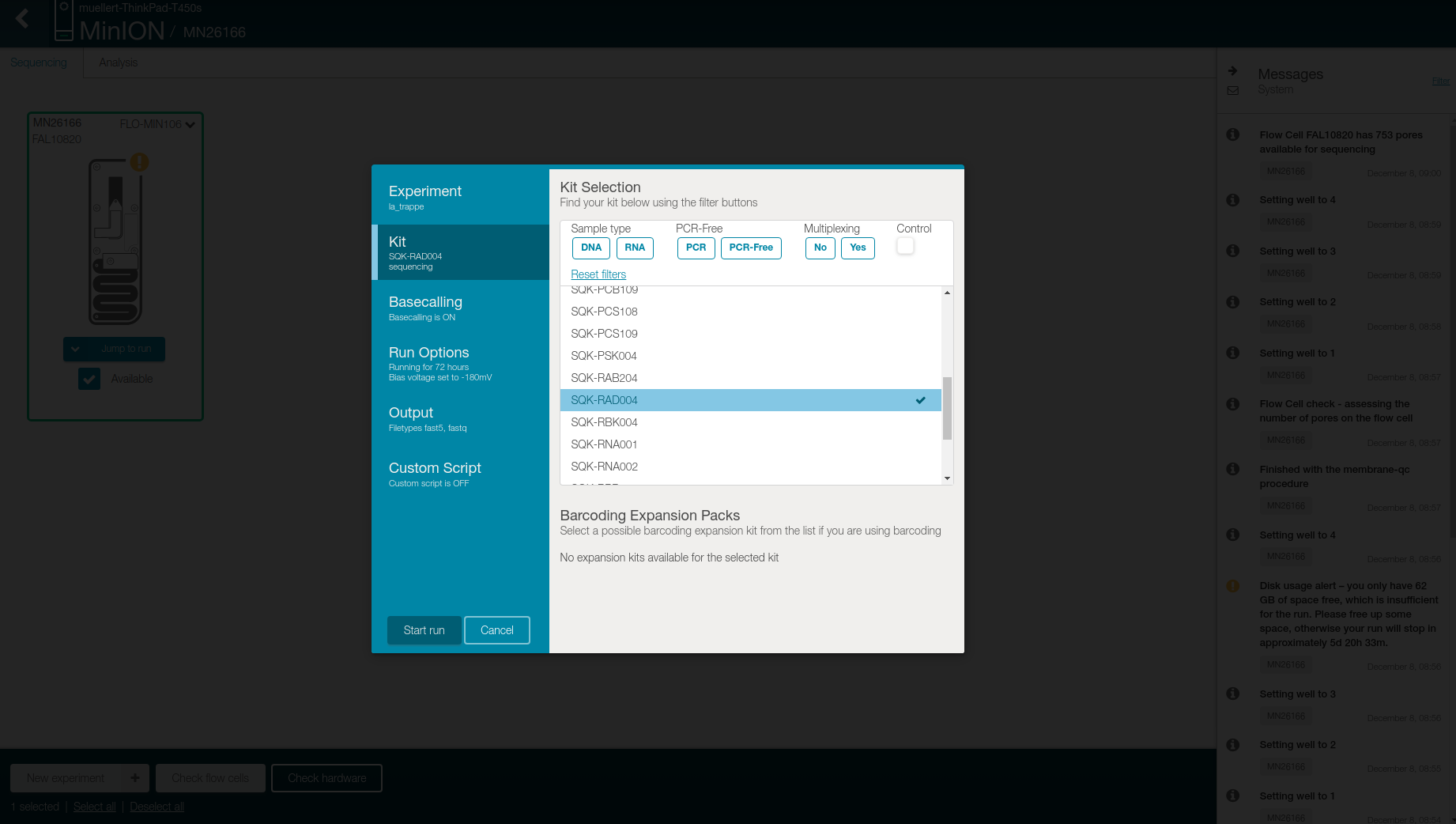
Select SQK-RAD004 in the Kit tab
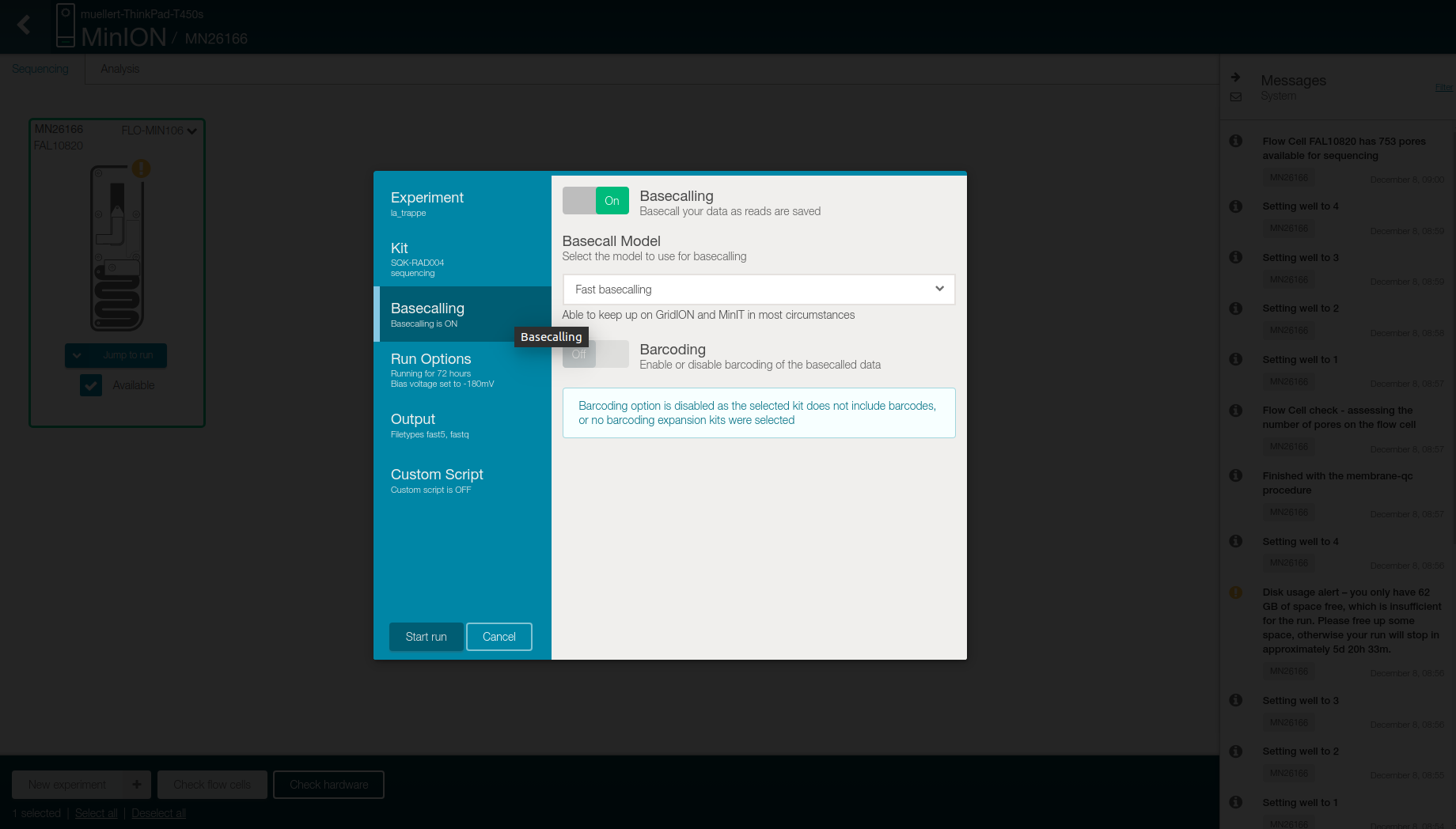
- Check in the Basecalling tab that
- “Basecalling” is “On”
- “Basecalling model” is “Fast basecalling”
- “Barcoding” is “Off”
-
Change the “Run lenght” to 4 in the Run Options tab
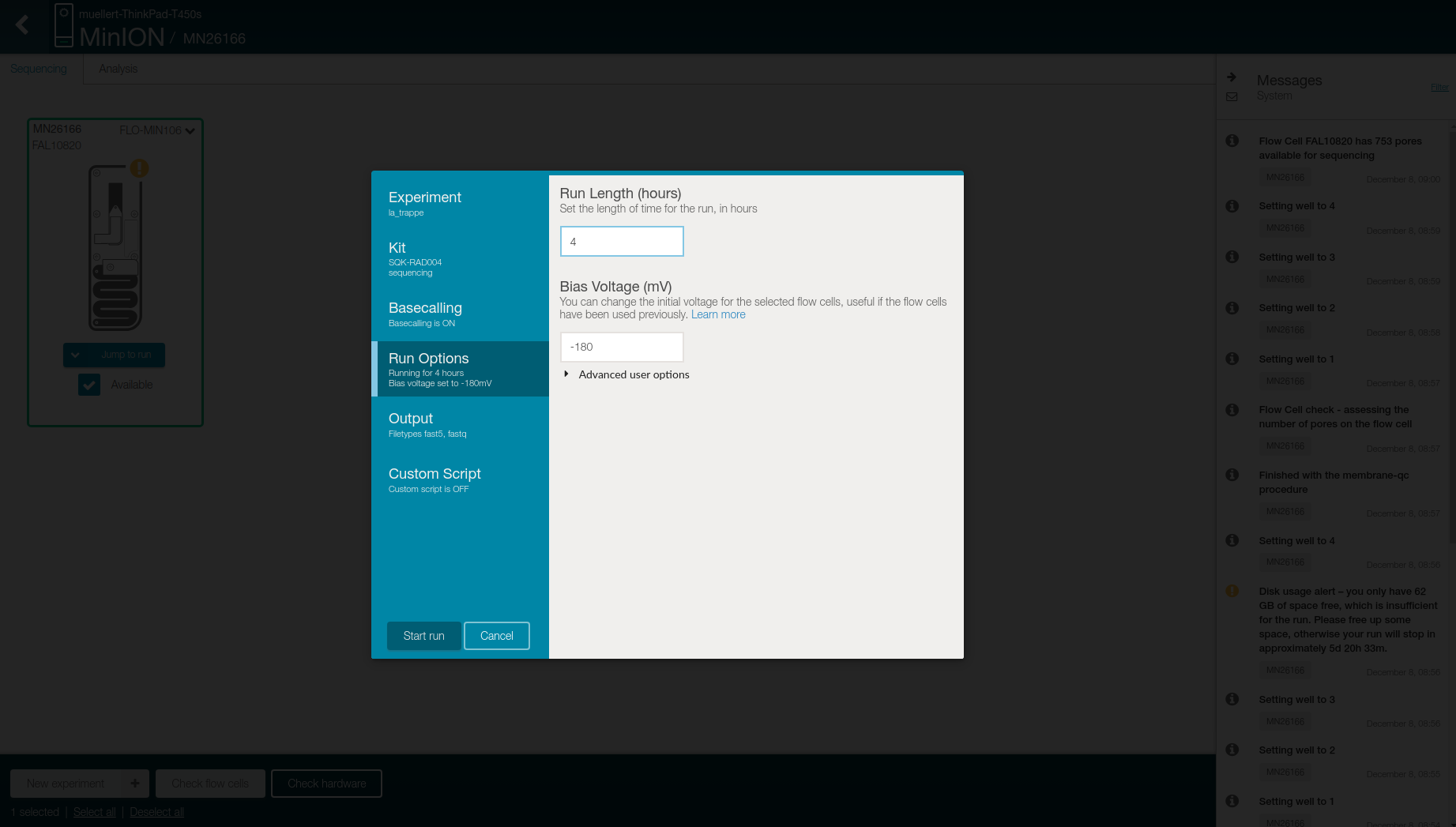
- Select in the Output tab
- Location to store the generated files
- Output format
- FAST5: raw signal, FASTQ record, and 4,000 reads per files
- FASTQ: 4,000 reads per files
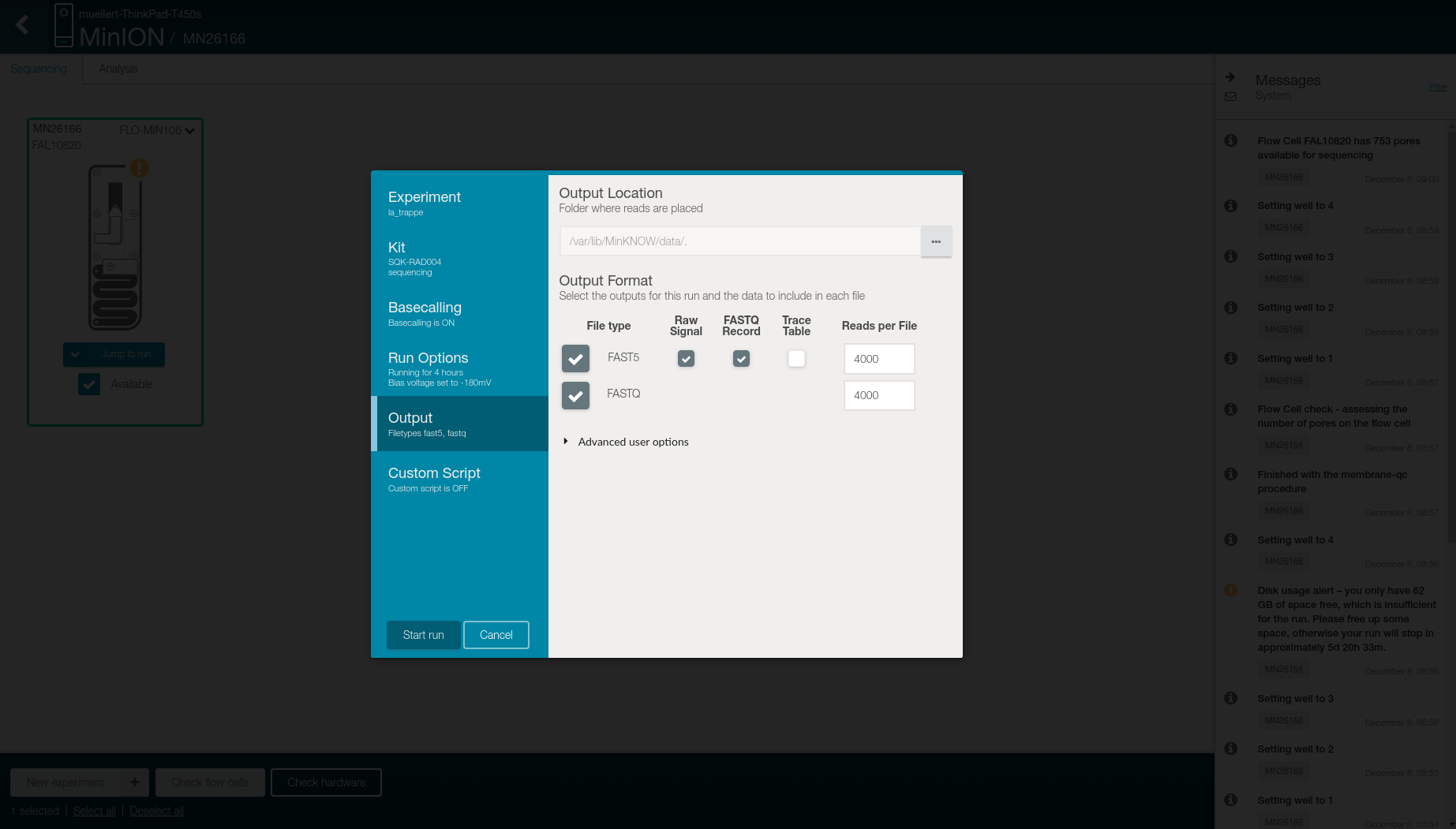
- Click on Start run
Once the sequencing started, information will be available to follow the sequencing process:
-
Status of the nanopore (1st graph)
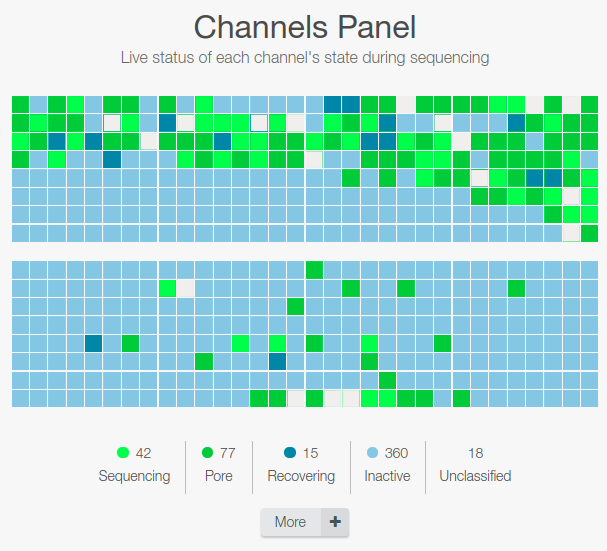
Check that the number of active pores reported are similar (within 10-15%) to those reported at the end of the Flow Cell Check
-
Number of DNA fragments (reads) that have been sequenced
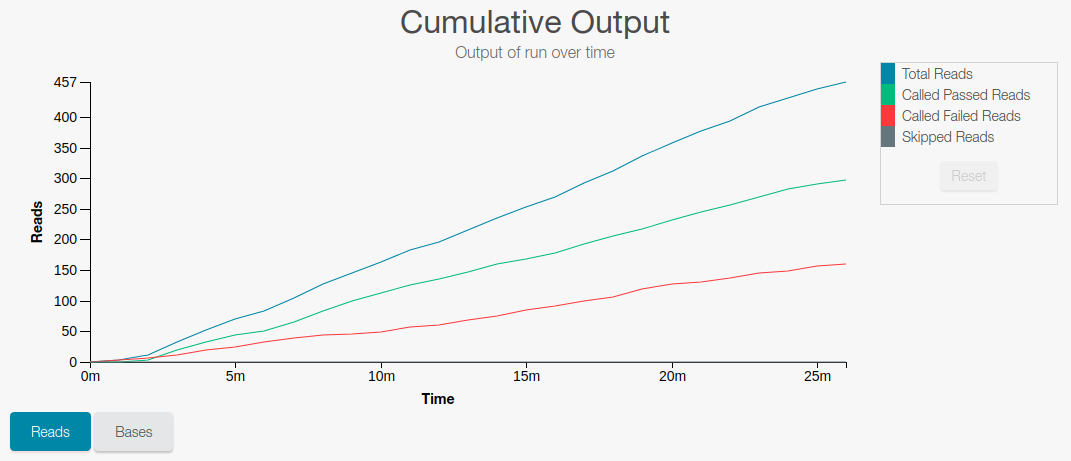
-
Length (number of bases) sequenced for the DNA fragments
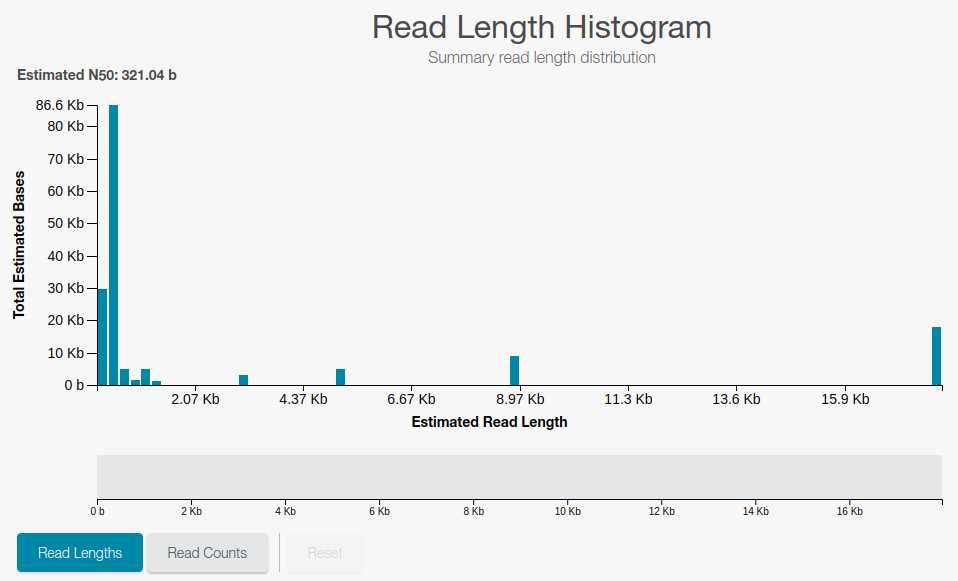
Cleaning
- Quit MinKNOW by closing it down
- Disconnect the MinION
- Wash the flow cell with the Washing Kit
- Remove the flow cell from the MinION
- Put the Configuration Test Cell back in