Purpose: Introduce biology and genomic science to the public and make pupils and citizens aware about DNA, sequencing technologies, bioinformatics, open science and their applications and their impact on their daily life
Process: 1-2 days continuous (or divided over several days) practical workshop on beer decoding
Hands-on workshop
Targets: pupils in high school (16 years old or older) and citizens with or without scientific background
Learning objectives: at the end of the workshop, participants will be able to:
- perform simple laboratory tasks
- give examples of microbes and their diverse roles
- explain what is cell and what it contains
- describe DNA as the building block for life
- give examples of DNA sequencing technologies
- apply a bioinformatics pipeline
Available pedagogical material
- Online, free to use and reuse
- Ready to use and modular protocols, with designated versions for teachers and pupil, and pedagogical gamification
- Presentation material to introduce the topic, theoretical background for lab-work and bioinformatics, with simple vocabulary
- Handbook to run a workshop independently (requirements and suggested timeline)
3 ways to run a workshop
- At the university campus with us
- On site (e.g. at your school) with us
- Self-organize using the material online
Previous workshops
We already organized several BeerDEcoded workshops over the last year. You can read more about them by checking our events.
Why?
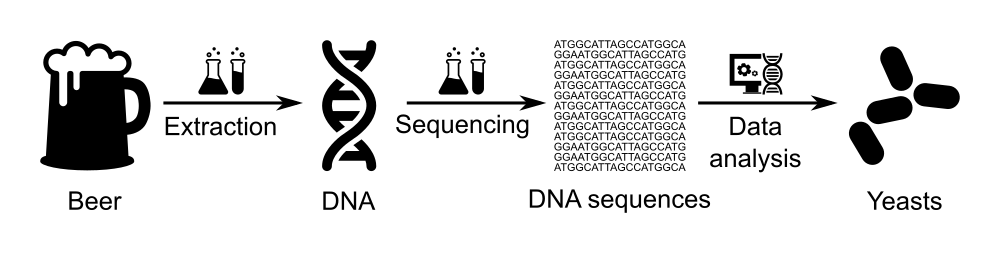
Beer is alive! It contains DNA that comes from its ingredients (hops, malts, yeast) and some hundred microbes. There are 1,000+ yeasts used for brewing and 200+ hop varieties, each one bearing a different DNA and contributing to differentiate its properties.
We can use the beer to sensibilize the public to molecular biology and genomic related research, DNA sequencing and data analysis and the challenges and possibilities that genomics brings to our society.
To bring the public to the contact of molecular biology, data-analysis, and open science, we organize workshops in which pupils, students and citizens extract, sequence and analyze the “DNA of beer”.
How?
This project was started by Hackuarium, an open Laboratory for DIY Biology, in Lausanne.
In Freiburg, we develop a prototype of the DNA extraction, sequencing, and analysis protocols using the Nanopore MinION, a portable real-time device for DNA sequencing, and Galaxy, an open source, web-based platform for computational biology and bioinformatics.
