Today, we had an unofficial kickoff of the BeerDEcoded project. We poured Beer into the MinION’s pores: sequencing yeast extracted from Beer with Nanopore.
During this day, we reproduced and improved the DNA extraction protocol that Stephan has developed. We also used for the 1st time our MinION. The results are very promising and the initial feedback is overwhelming. It was a blast to do some wet-lab work, specially as most of us, as bioinformaticians and computer scientists, are not used to it!





DNA extraction
We used a Chimay red and wanted to extract the DNA of the yeast inside this beer. To do so, we:
- Extracted the yeast from the beer with several centrifuge runs


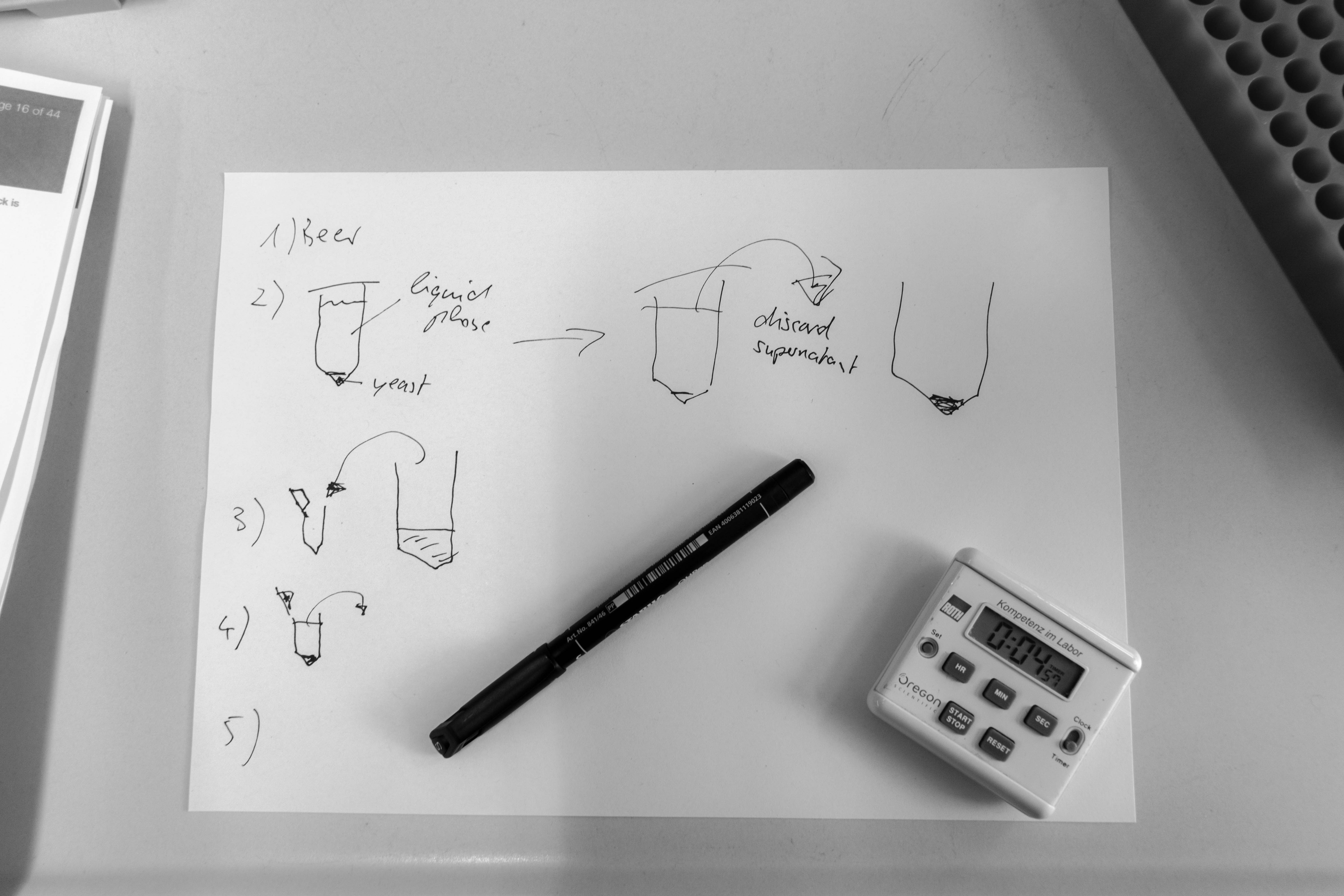
- Broke the cell membranes of the yeasts using an ethanol bath and a lysis buffer
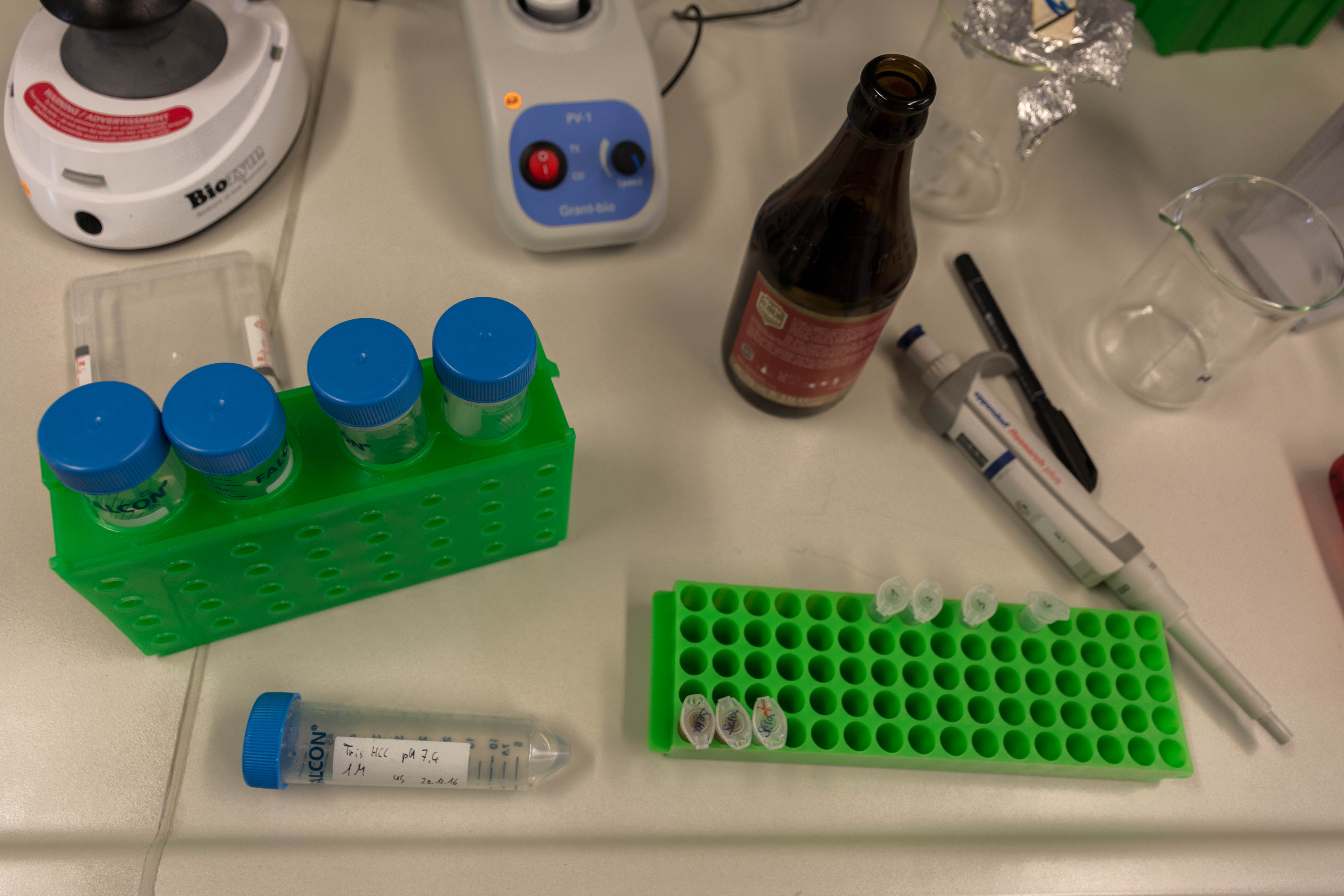


- Extracted the DNA from the cell using magnetic beads and a MinElute Reaction Kit 50 from Qiagen








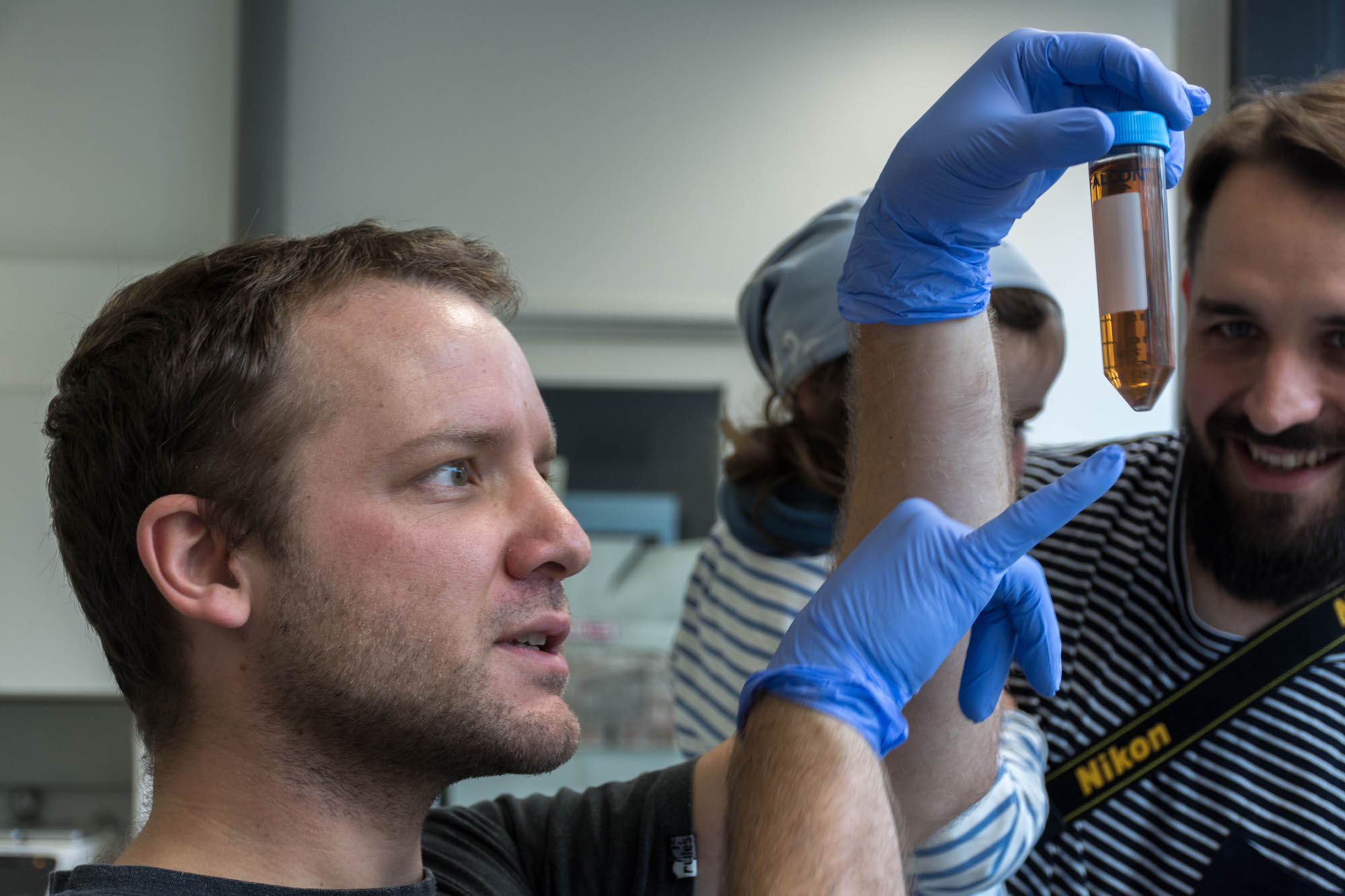
At the end we quickly check the purity with a spectrophotometer and freezed the DNA until library preparation.
The details of the extraction protocol we used can be found here.
DNA sequencing
During the DNA extracted, we started to play with the MinION with the installation of the Nanopore software and ran a functional test of the flowcell. We then did the library preparation: preparation of the DNA for sequencing.



We then loaded the MinION with our DNA and launched the sequencing



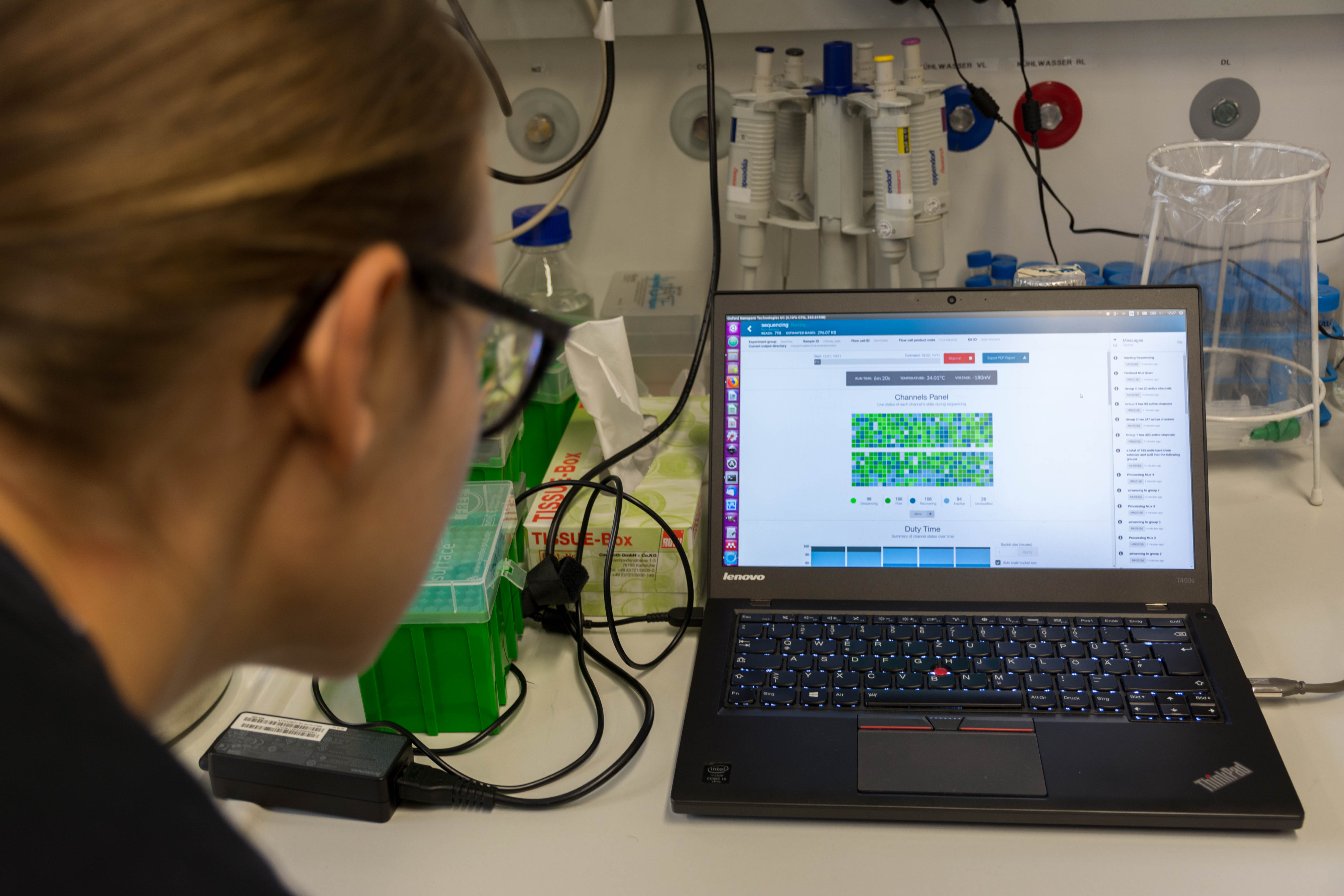
It was quite exciting. We had long reads until 24kb, but the quality started to decrease quite quickly (probably not enough DNA) so we stopped it early after our lunch break.
The details of the sequencing protocol we used can be found here.
Milad then analyzed the generated sequences and extracted some Saccharomyces cerevisae.
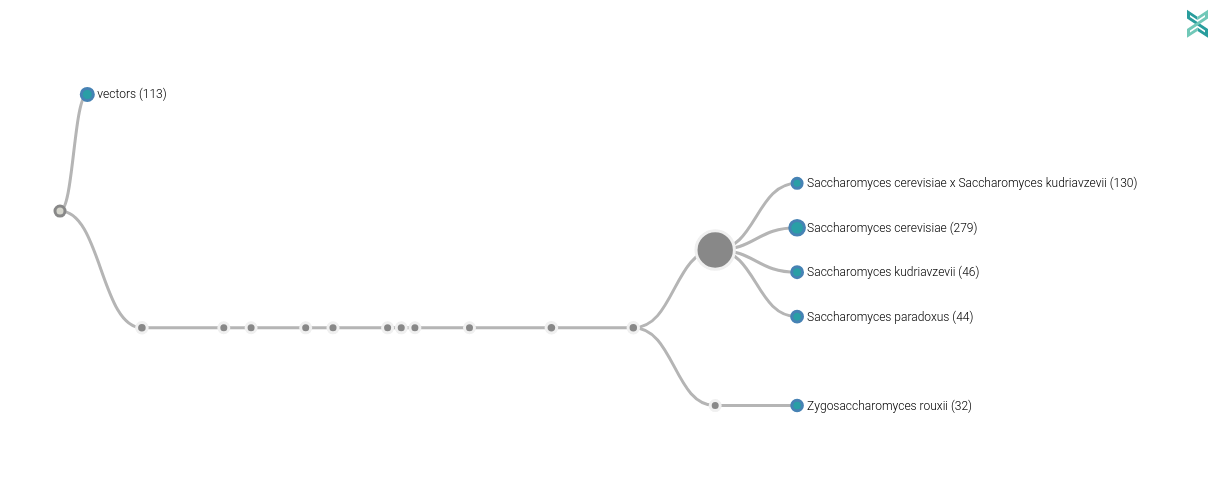
Very promising results (given we stoped the sequencing)!
Next steps
Our next challenge will be to simplify the DNA extraction protocol, specifically using simpler and everyday life tools, and ensure its robustness on every location and situation, i.e., the protocol has to work in a specialised lab as well as in a high school chemistry/biology labs. To overcome this challenge, we will collect ideas from Do-It-Yourself biology projects.
We will make special efforts in making the protocols accessible and understandable by everyone, with graphics supporting each step and explaining their importance and their execution.
For the data analysis, a dedicated workflow will be available via Galaxy. This open source, web-based platform makes reproducible computational biology and bioinformatics accessible to anyone via a web-browser.
A dedicated Galaxy server (streetscience.usegalaxy.eu) will be implemented to serve the workflow. In complement, training material for data analysis will be created in cooperation with the Galaxy Training community with slides, hands-on material, interactive web-tours suitable to guide our participants from data upload to data analysis.
As the aim of this project is to share our knowledge with the public, we will organize two-day “DNA extraction, sequencing and data analysis” workshops, one at a local biotechnology high school, and one for a larger audience.
Check out more on Twitter
The pictures by Bérénice Batut are licensed under CC-BY-SA.